Title page
期刊:PLoS One
年份:2021 Aug 17
数据集链接:https://doi.org/10.7910/DVN/FCBUOR
pdf链接:
Summary
-
In this paper, we create an endoscopic dataset collected from various sources and annotate the ground truth of polyp location and classification results with the help of experienced gastroenterologists.
-
The dataset can serve as a benchmark platform to train and evaluate the machine learning models for polyp classification
-
We have also compared the performance of eight state-ofthe-art deep learning-based object detection models.
Workflow
无
Methods
Evaluation models for detection and classification
-
YOLOv3
-
YOLOv4
-
SSD
-
RetinaNet: a one-stage framework based on the SSD model, using the FPN and Focal loss
-
DetNet
- RefineDet
Dataset build
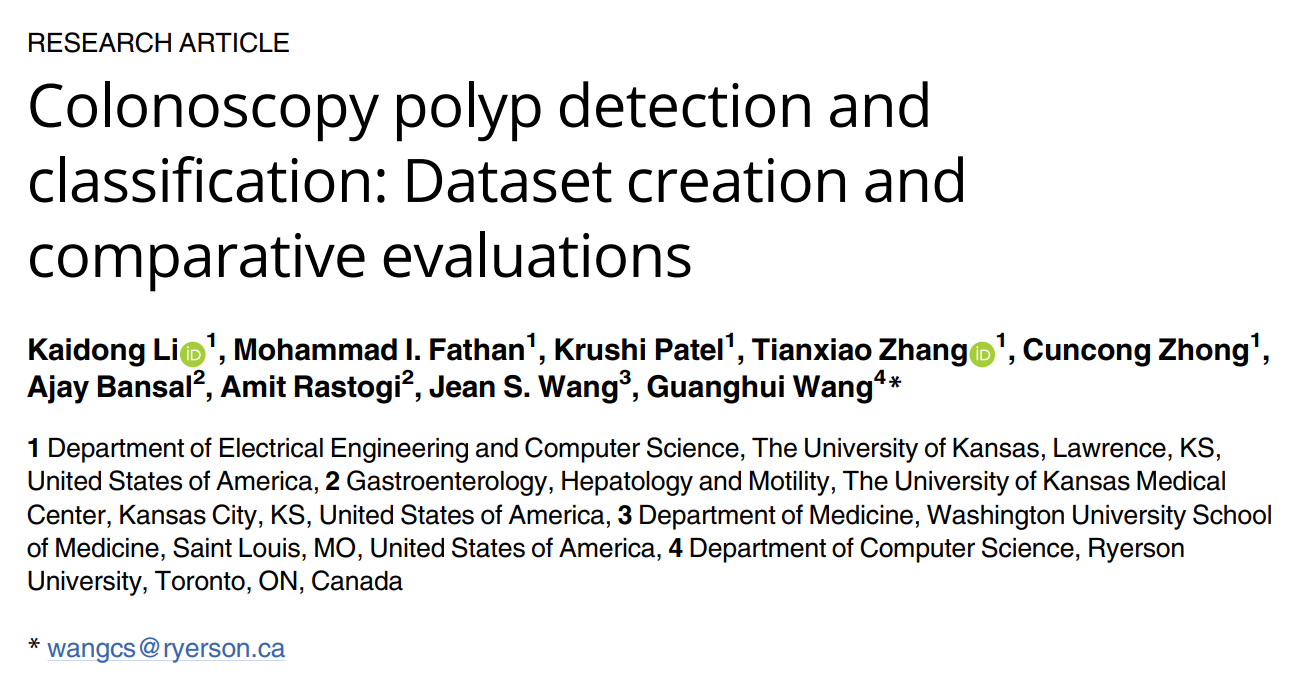
Images in different datasets vary greatly
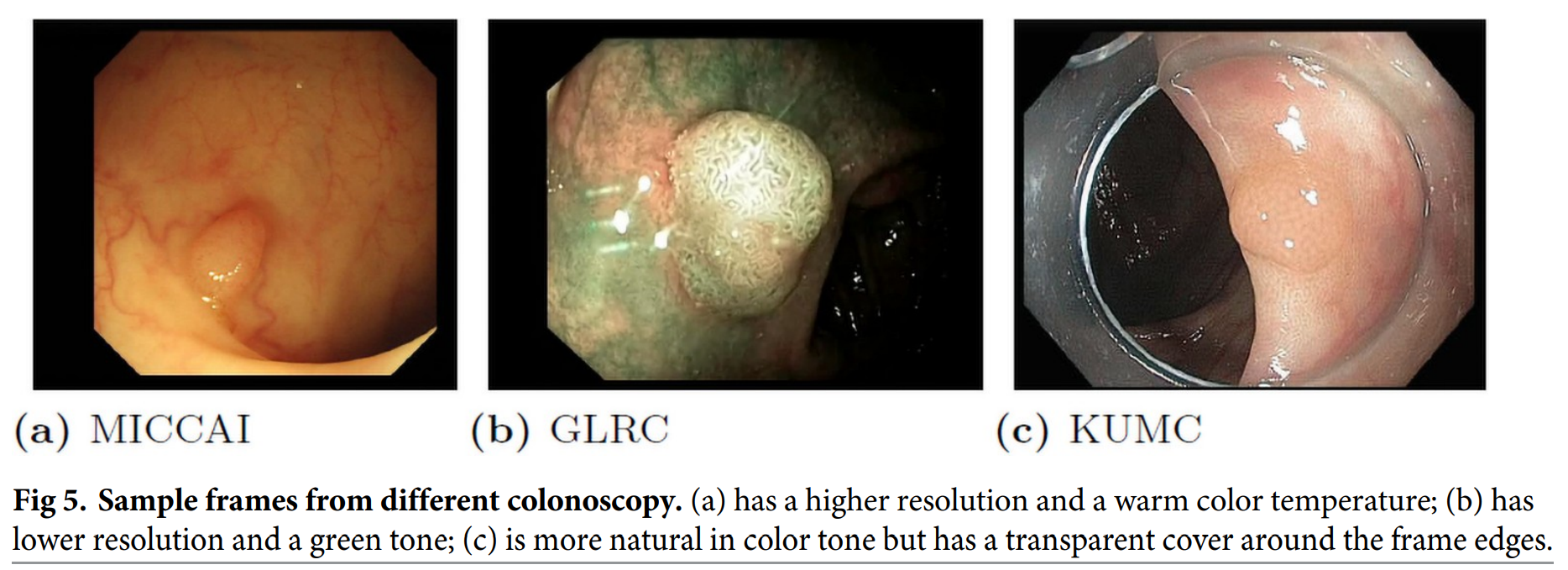
Each endoscopic video sequence has significant redundancies
Datasets selection and annotation
将所有数据集规整化为 bounding box + class(二分类:腺瘤/增生性息肉)
源数据:
MICCAI 2017:
- 18 videos for training and 20 videos for testing
- 只有掩码
CVC colon DB:
- 15 short colonoscopy videos with a total of 300 frames
- 只有掩码
GLRC dataset:
- 76 short video sequences with class labels
- 只有分类标签
KUMC dataset:
- 80 colonoscopy video sequences.
数据集筛选:
-
To avoid some long videos overwhelming others, we adopt an adaptive sampling rate to extract the frames from each video sequence based on the camera movement and video lengths
- After sampling, we extracted around 300 to 500 frames for long sequences to maintain a balance among different sequences, while for small sequences like CVC colon DB, we simply keep all image frames in the sequence.
- 分类标签:When the endoscopist could not reach an agreement on the classification results, we simply remove those sequences from the dataset.
数据集划分:
- We make the division for each dataset and polyp class independently
- For each class in one dataset, we randomly select 75%, 10%, and 15% sequences to form the training, validation, and test sets, respectively
Result-show
1. 数据集概览
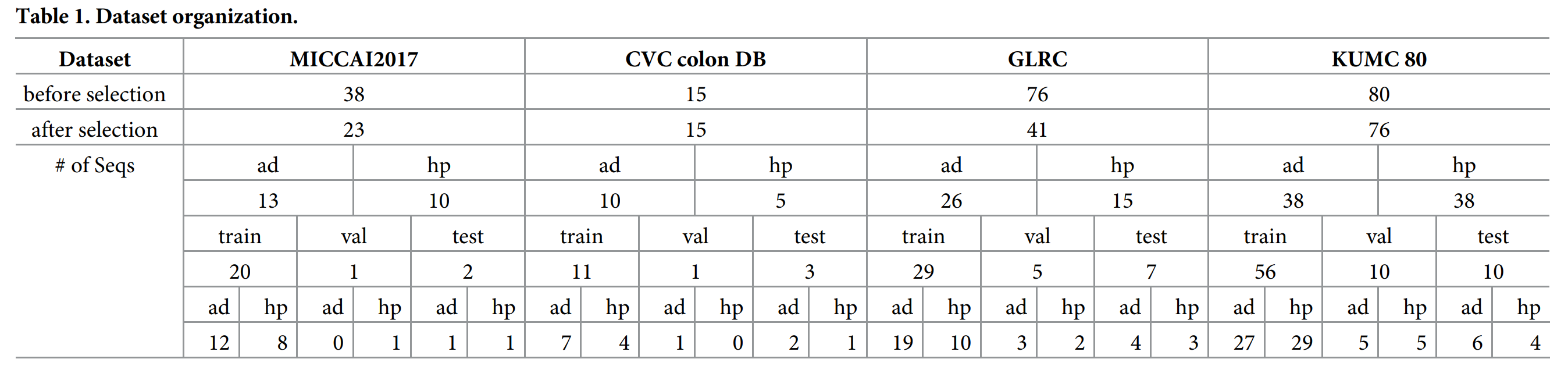
-
标注类型:bbox + 二分类结果(腺瘤/增生性息肉)
-
肠镜类型:待核查
-
116 training, 17 validation, and 22 test sequences
-
训练集 28773, 验证集4254, 测试集 4872 帧
3. 基准模型测试结果
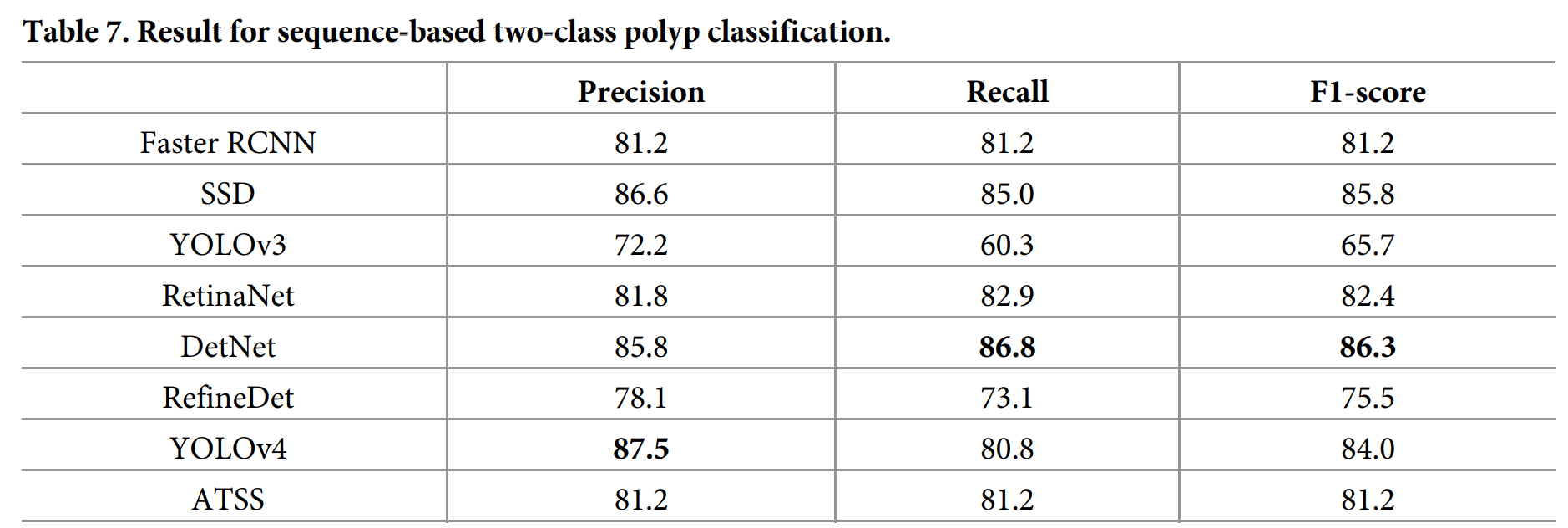
息肉分类结果
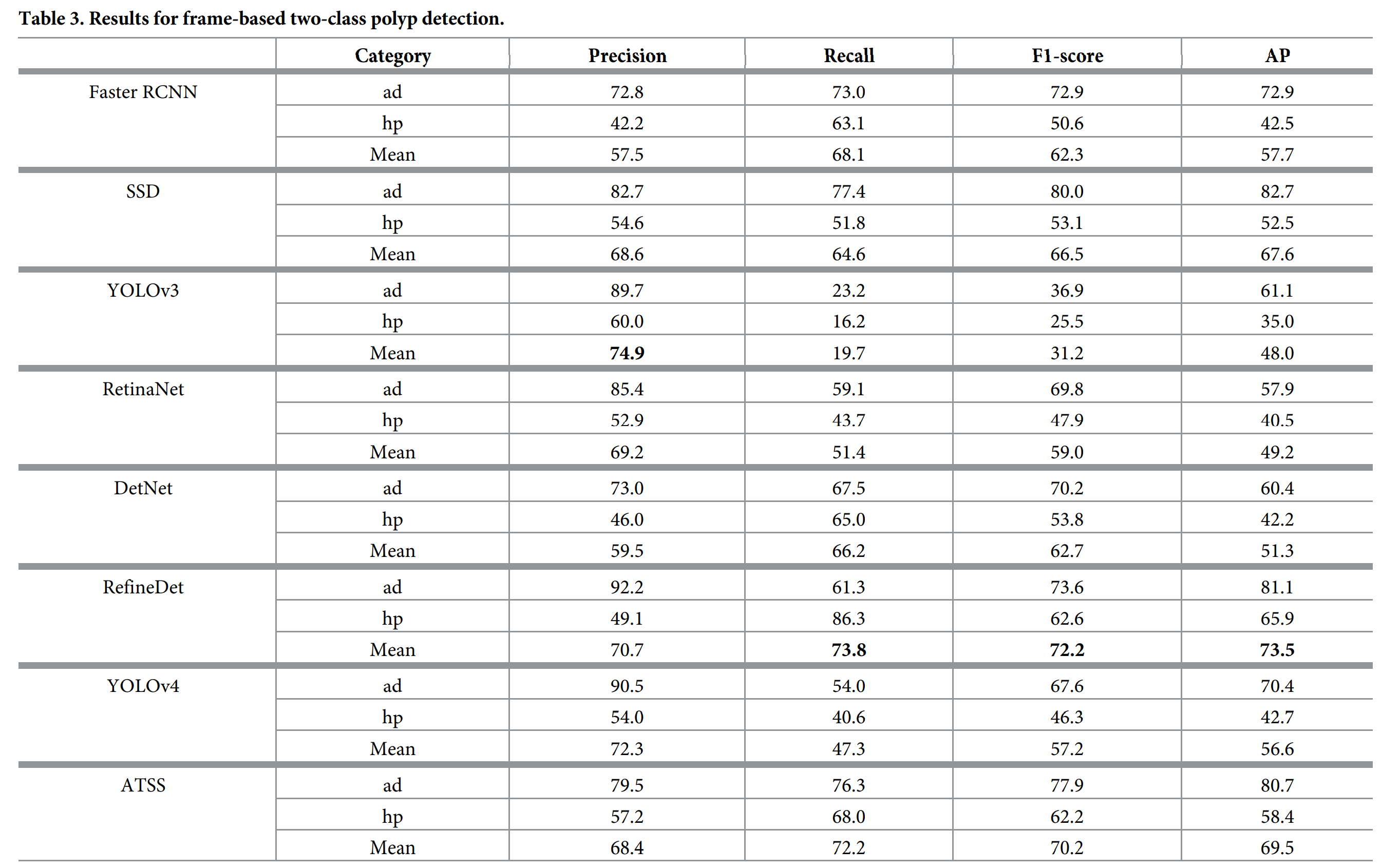
仅息肉检测的结果:
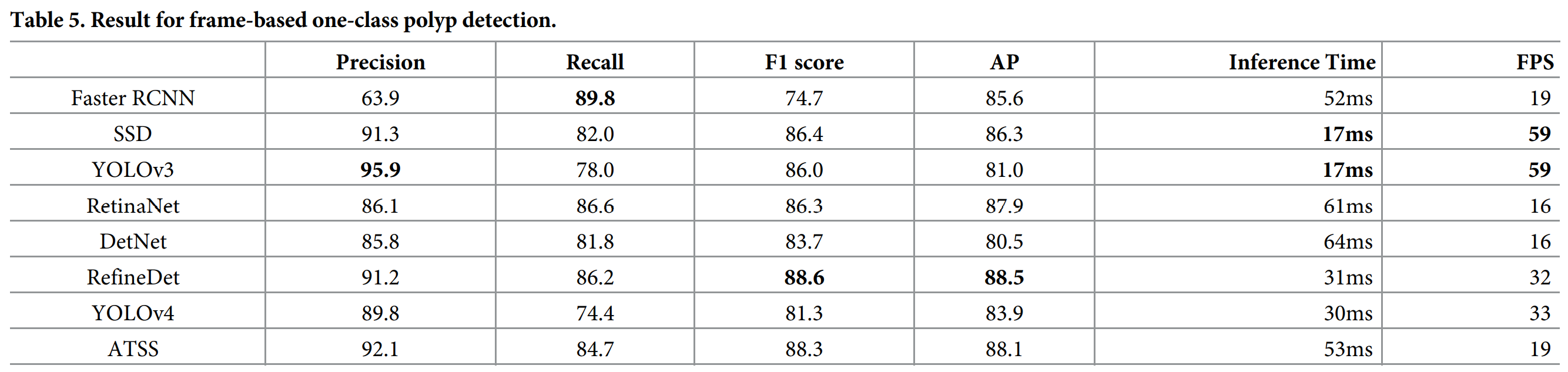
基于序列分析结果
数据集注释内容
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
# 数据集名称:
PolypSet
# 数据集结构
--train2019
Annotation
1.xml (28773个文件)
2.xml
...
Image
1.jpg (28773个文件)
2.jpg
...
--test2019
- image
- 1 (22个文件夹)
1.jpg
2.jpg
...
- 2
- ...
- Annotation
- 1 (22个文件夹)
1.xml
2.xml
...
- 2
- 3
--val2019
- 1(17个文件夹)
- image
1.jpg
2.jpg
3.jpg
...
- Annotation
1.xml
2.xml
3.xml
...
- 2
- ...
# Annotation example:
# 1.xml
<annotation>
<folder>3</folder>
<filename>245.png</filename>
<path>/scratch/mfathan/Thesis/Dataset/Extracted/MICCAI2017_Test/test/3/245.png</path>
<source>
<database>Unknown</database>
</source>
<size>
<width>384</width>
<height>288</height>
<depth>3</depth>
</size>
<segmented>0</segmented>
<object>
<name>adenomatous</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>164</xmin>
<ymin> 113</ymin>
<xmax> 343</xmax>
<ymax> 279</ymax>
</bndbox>
</object>
</annotation>