Title page
会议:Accepted at IPMI 2021
年份:2021
github链接:https://github.com/zhangybzbo/ColonLight
pdf链接:https://arxiv.org/pdf/2103.10310.pdf
Summary
- The lighting inconsistency of colonoscopy videos can cause a key component of the colonoscopic reconstruction system
- 这篇文章找到了一种光线校正的方法,来调节邻近视频帧之间的光线密度分布
- 该方法使用RNN网络来无监督地调整gamma值,实现了实时
- 该方法显著增加了肠镜表面重建系统的重建成功率与重建质量
Workflow
Methods
1. SLAM mechanism
-
Simultaneous Localization And Mapping (SLAM), one of the most successful methods for 3D reconstruction.
-
SLAM is an algorithm that can achieve real-time dense reconstruction from a sequence of monocular images
-
SLAM has a localization component and a mapping component; the two components operate cooperatively. The localization (tracking) component predicts the camera poses from each incoming image frame. Based on the visual clues extracted from the images, the mapping component optimizes especially the pose predictions but also the keypoints’ depth estimates
-
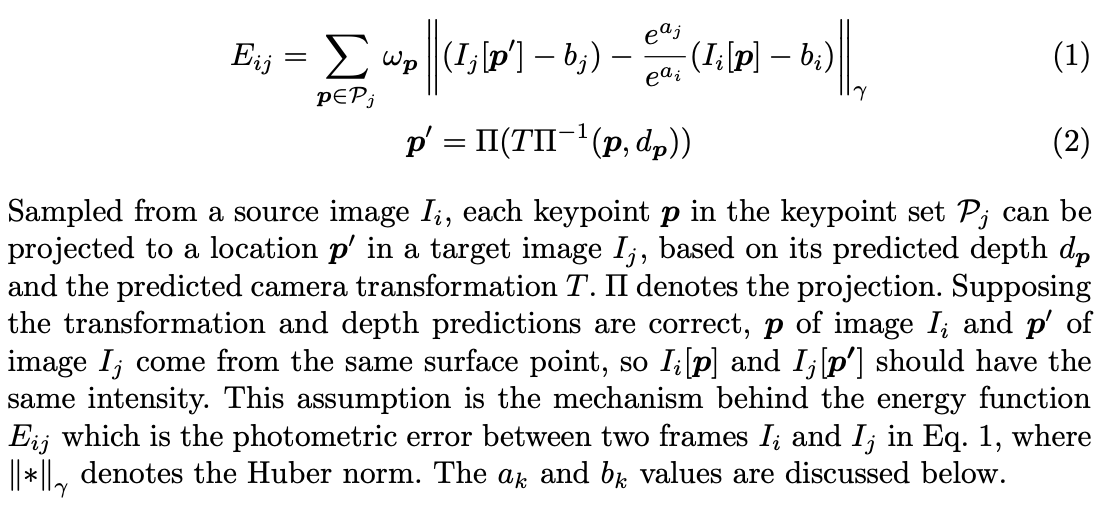
目标函数:
The lighting problem in colonoscopic surface reconstruction
- 肠镜的特殊之处:the point light is moving with the camera and can change rapidly due to motion and occlusion
- SLAM的额外亮度优化不能很好地处理肠镜点光源的问题,such as contrast difference (bright regions become brighter and dark regions become darker).
In this work we apply an adaptive intensity mapping to enhance the colonoscopy frame sequence with the help of an RNN network。
实现细节
1. RNN → gamma值调整(实时考虑)
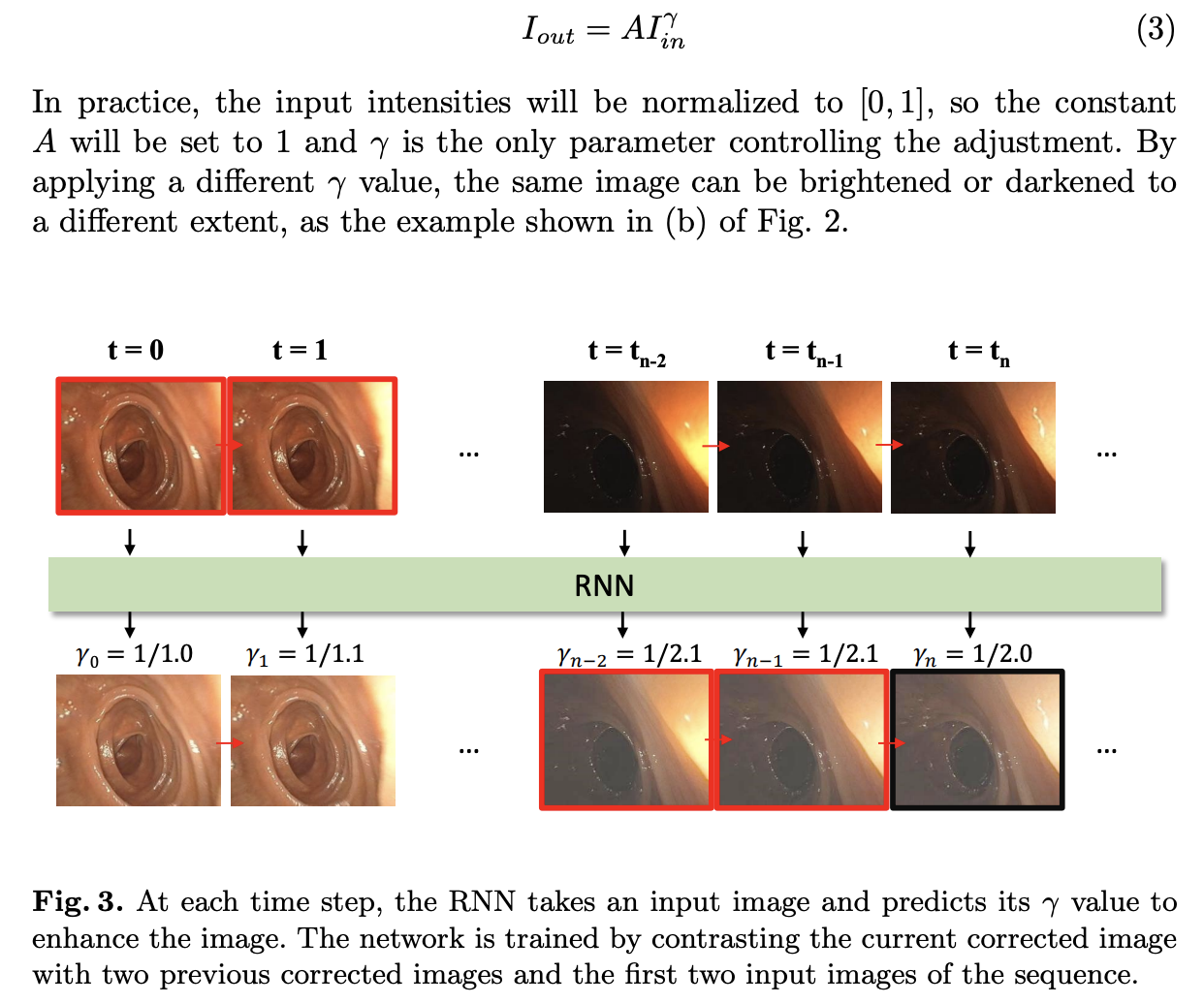
2. RNN Network
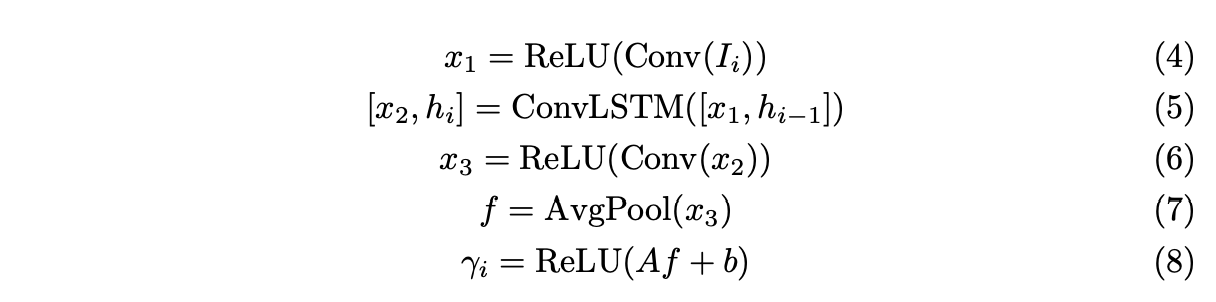
3. 训练方式:无监督
-
使用输入图像的前两帧与整个视频序列(共10帧)的前两帧作为参考
-
损失函数:结构相似性测量
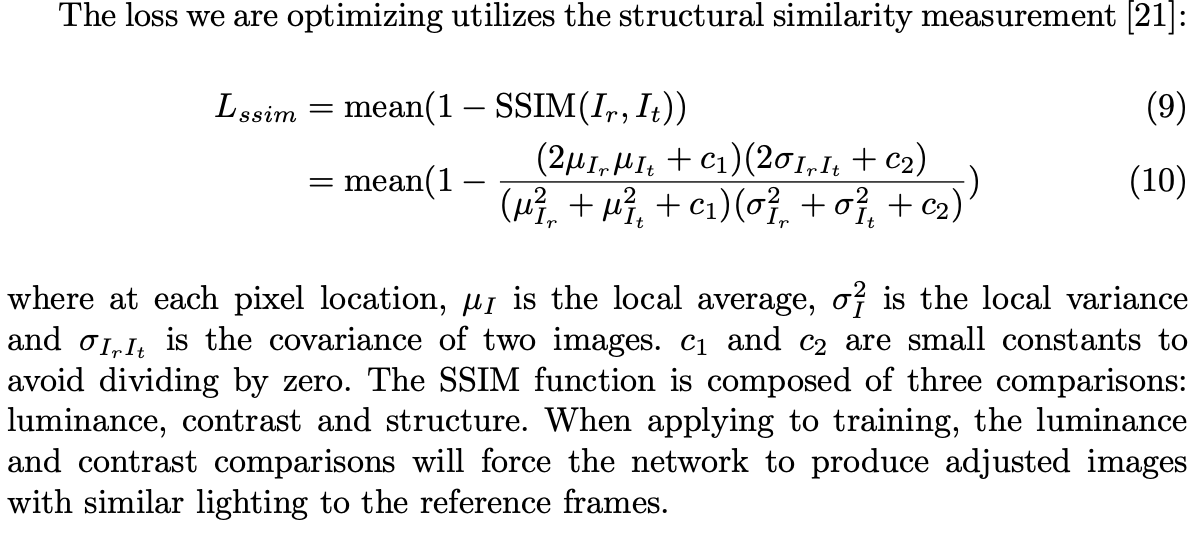
-
In training when computing the Lssim, we mask out the pixels with input intensity larger than 0.7
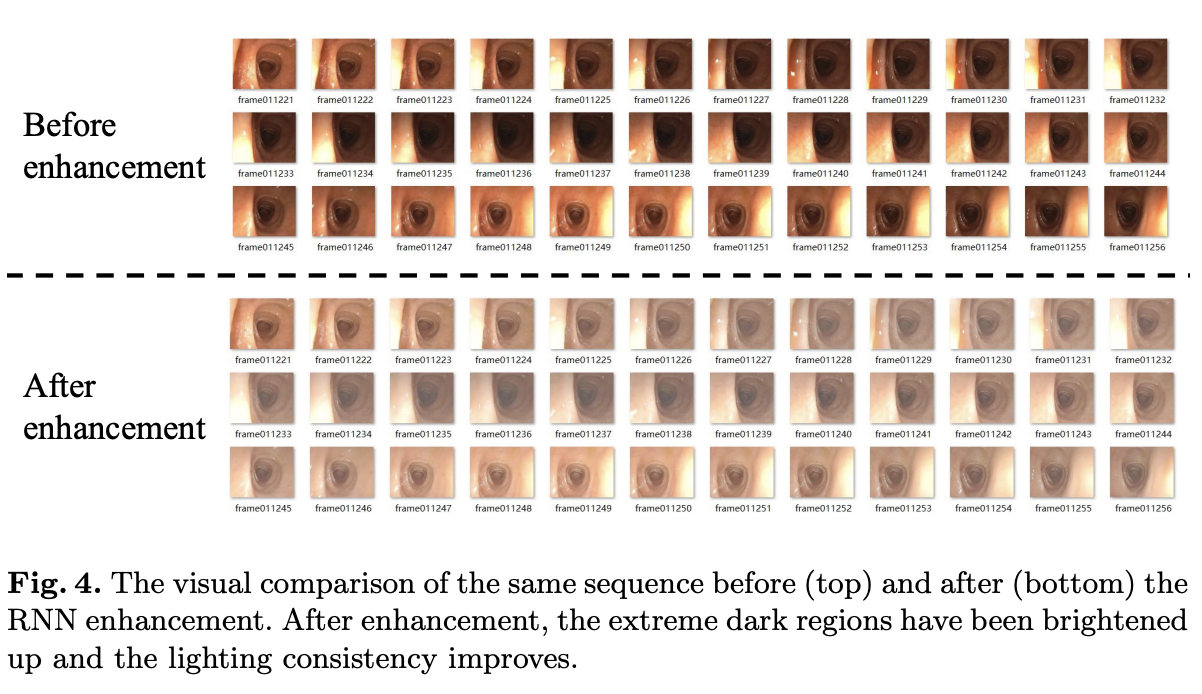
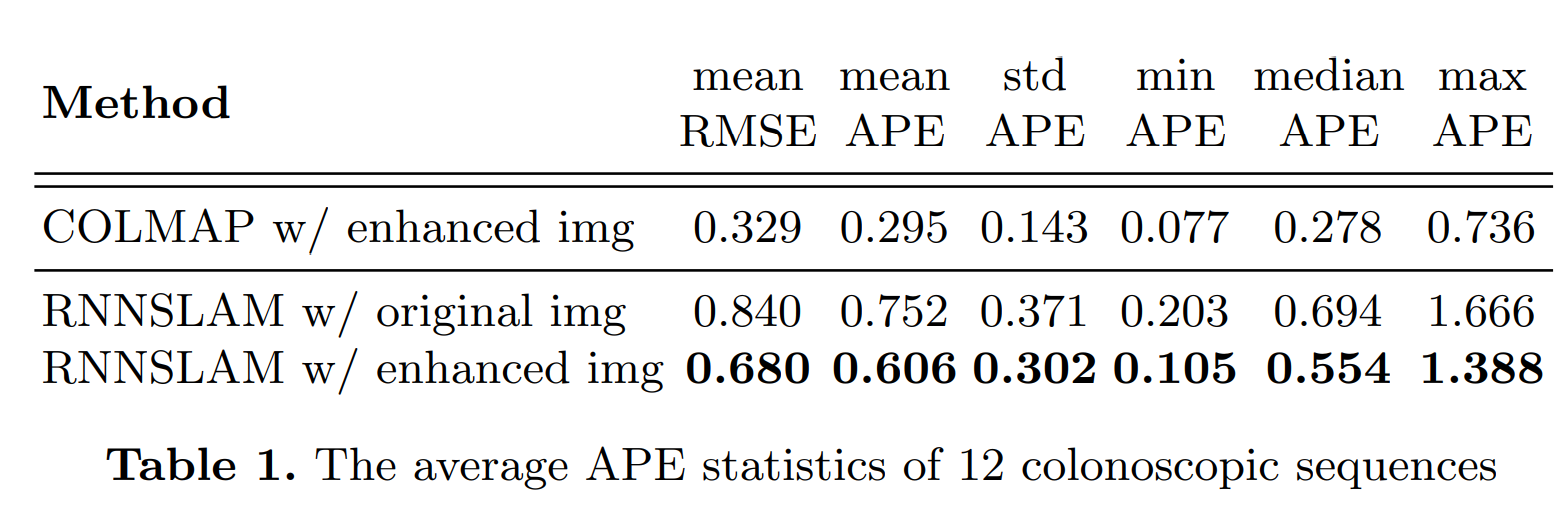
Result-show
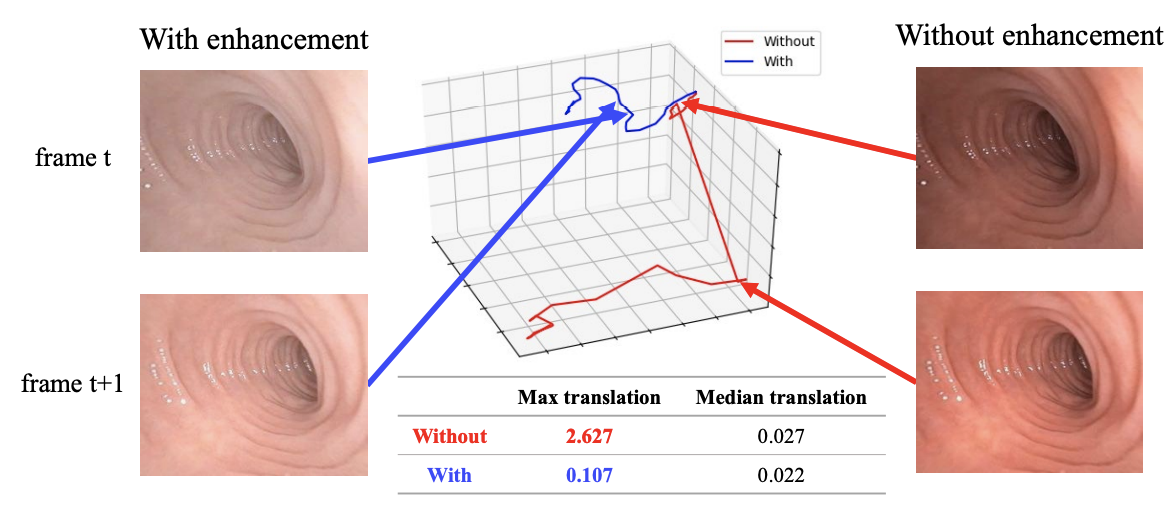
APE:Absolute pose error
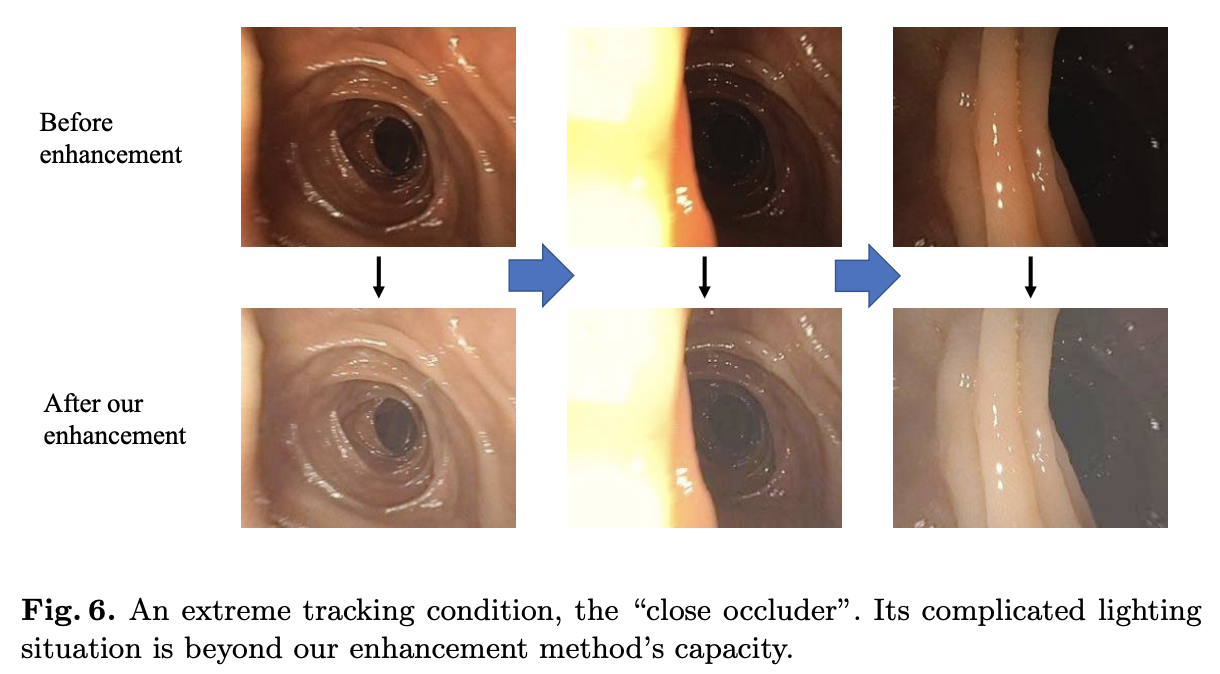
启发和思考
- 可否借鉴,用于息肉分割的阈值后处理
- 可否用于前处理,通过调整gamma值与其他值,结合域自适应,使得输入的肠镜图像能够快速归一化
代码注释
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
import torch
import torch.nn as nn
import torch.nn.functional as F
'''modified from https://github.com/ndrplz/ConvLSTM_pytorch/'''
class ConvLSTMCell(nn.Module):
def __init__(self, input_dim, hidden_dim, kernel_size, bias=True, forget_bias=1.0):
"""
Initialize ConvLSTM cell.
Parameters
----------
input_dim: int
Number of channels of input tensor.
hidden_dim: int
Number of channels of hidden state.
kernel_size: (int, int)
Size of the convolutional kernel.
bias: bool
Whether or not to add the bias.
"""
super(ConvLSTMCell, self).__init__()
self.input_dim = input_dim
self.hidden_dim = hidden_dim
self.kernel_size = kernel_size
self.padding = kernel_size[0] // 2, kernel_size[1] // 2
self.bias = bias
self.forget_bias = forget_bias # TODO: add or not
self.conv = nn.Conv2d(in_channels=self.input_dim + self.hidden_dim,
out_channels=4 * self.hidden_dim,
kernel_size=self.kernel_size,
padding=self.padding,
bias=self.bias)
torch.nn.init.xavier_normal_(self.conv.weight)
def forward(self, input_tensor, cur_state):
'''
:param input_tensor: B x input_dim x height x width
:param cur_state: B x 2hidden_dim x height x width
:return: h_next, [h_next, c_next]
'''
h_cur, c_cur = torch.split(cur_state, self.hidden_dim, dim=1)
combined = torch.cat([input_tensor, h_cur], dim=1) # concatenate along channel axis
combined_conv = self.conv(combined)
cc_i, cc_f, cc_o, cc_g = torch.split(combined_conv, self.hidden_dim, dim=1)
i = torch.sigmoid(cc_i)
f = torch.sigmoid(cc_f + self.forget_bias)
o = torch.sigmoid(cc_o)
g = torch.tanh(cc_g)
c_next = f * c_cur + i * g
h_next = o * torch.tanh(c_next)
return h_next, torch.cat([h_next, c_next], dim=1)
def init_hidden(self, batch_size, image_size):
height, width = image_size
return (torch.zeros(batch_size, self.hidden_dim, height, width, device=self.conv.weight.device),
torch.zeros(batch_size, self.hidden_dim, height, width, device=self.conv.weight.device))
class RNNmodule(nn.Module):
def __init__(self, img_channel, hidden_channel, adj_channel, kernel_size, pixelwise):
super(RNNmodule, self).__init__()
self.img_channel = img_channel
self.adj_channel = adj_channel
self.pixelwise = pixelwise
self.padding = kernel_size[0] // 2, kernel_size[1] // 2
self.in_cnv = nn.Conv2d(img_channel, hidden_channel, kernel_size, stride=1, padding=self.padding)
self.bn = nn.BatchNorm2d(hidden_channel, momentum=0.01) # TODO
self.RNNcell = ConvLSTMCell(hidden_channel, hidden_channel, kernel_size)
self.rnn_cnv = nn.Conv2d(hidden_channel, hidden_channel, kernel_size, stride=1, padding=self.padding)
if pixelwise:
self.out_cnv = nn.Conv2d(hidden_channel, adj_channel, kernel_size, stride=1, padding=self.padding)
else:
self.out_linear = nn.Linear(hidden_channel, 1)
# TODO: or kernel size=1
# TODO: or multi-adjustment using rnn
self.init_param()
def init_param(self):
torch.nn.init.xavier_normal_(self.in_cnv.weight)
torch.nn.init.xavier_normal_(self.rnn_cnv.weight)
if self.pixelwise:
torch.nn.init.xavier_normal_(self.out_cnv.weight)
self.bn.weight.data.normal_(1.0, 0.02)
self.bn.bias.data.fill_(0)
def forward(self, inputs, last_hidden):
b, c, h, w = inputs.size()
assert c == self.img_channel
if last_hidden == None:
last_hidden = torch.cat(self.RNNcell.init_hidden(b, (h, w)), dim=1)
conv = self.in_cnv(inputs)
conv = self.bn(conv)
conv = F.relu(conv)
rnn, new_hidden = self.RNNcell(conv, last_hidden)
rnn = self.rnn_cnv(rnn)
# rnn = self.bn(rnn)
rnn = F.relu(rnn)
if self.pixelwise:
adjs = F.relu(self.out_cnv(rnn))
adj_t = torch.split(adjs, 1, dim=1)
x = inputs
for i in range(self.adj_channel):
x = torch.pow(x, 1/adj_t[i]) # adj_t[i] B x 1 x h x w
else:
adjs = torch.mean(rnn, dim=[-1, -2])
adjs = F.relu(self.out_linear(adjs)) # B x 1
adjs = adjs.view(-1, 1, 1, 1)
x = torch.pow(inputs, 1/adjs)
# x_gray = torch.mean(x, dim=1, keepdim=True)
# xp = adjs[:, 0].view(-1, 1, 1, 1)
# yp = adjs[:, 1].view(-1, 1, 1, 1)
# x = (x_gray<=xp) * yp/xp * x + (x_gray>xp) * ((1-yp)/(1-xp) * x + (yp-xp)/(1-xp))
return x, adjs,